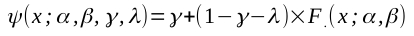I am not sure I understand some details in the original post ( e.g., why send data into PAL_CumulativeNormal using the 'inverse' argument?). But I think I get the gist of the issue. It might indeed be a good idea to refit data obtained in a PAL_AMPM run in a separate fit. The estimates given in the psi object [e.g., PM.threshold(end)] are based on the posterior distribution on the grid that you defined (using 'priorAlphaRange', etc. arguments). In the psi method, this grid defined over a finite domain and at a limited granularity. If your posterior runs off of this domain, this would influence your estimate. Also, if your granularity is very coarse, this will influence your estimate. Refitting with PAL_PFHB_modelFit gets around these issues, but there would still be differences: the estimates in the psi objects are marginal means of the posterior, whereas PAL_PFHB by default (but this can be changed) uses the marginal modes. Also, by default: PAL_PFHB uses a mildly informative prior for the lapse rate, whereas the psi method uses a uniform prior. Refitting using PAL_PFML_Fit is a bit questionable to purists. PAL_AMPM optimizes with respect to a Bayesian estimate, but PAL_PFML_Fit uses a maximum likelihood criterion. So your optimization is not optimal. Nevertheless, in the end all estimates should all estimate the same quality (the true generating value) so if they do their job well they should all be similar, at least if a decent number of trials is used. If you can reproduce a case where estimates are very different you can post here and I can take a look. Ideally something I can reproduce exactly. E.g., I whipped up the code below. Code can also be downloaded
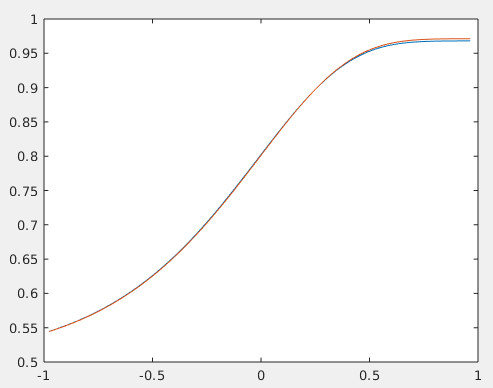
here. It runs a psi-method run, then refits the data using PAL_PFHB_Fit, then plots the PFs for each fit (which in this example are virtually identical, see image). They're not exactly the same for reason mentioned above. If you have code like this (but feel free to change anything including using PAL_PFML_Fit instead of PAL_PFHB_modelFit) where the fits are very different, please post here.
rng(0); %Include seed so it will reproduce exactly
NumTrials = 240;
grain = 201;
PF = @PAL_Gumbel; %assumed psychometric function
stimRange = (linspace(PF([0 1 0 0],.1,'inverse'),PF([0 1 0 0],.9999,'inverse'),21));
priorAlphaRange = linspace(PF([0 1 0 0],.1,'inverse'),PF([0 1 0 0],.9999,'inverse'),grain);
priorBetaRange = linspace(log10(.0625),log10(16),grain);
priorGammaRange = 0.5;
priorLambdaRange = 0:.01:.2;
%Initialize PM structure
PM = PAL_AMPM_setupPM('priorAlphaRange',priorAlphaRange,...
'priorBetaRange',priorBetaRange,...
'priorGammaRange',priorGammaRange,...
'priorLambdaRange',priorLambdaRange,...
'numtrials',NumTrials,...
'PF' , PF,...
'stimRange',stimRange);
paramsGen = [0, 1, .5, .02]; %generating parameter values [alpha, beta, gamma, lambda] (or [threshold, slope, guess, lapse])
while PM.stop ~= 1
response = rand(1) < PF(paramsGen, PM.xCurrent); %simulate observer
%update PM based on response:
PM = PAL_AMPM_updatePM(PM,response);
end
data.x = PM.x(1:end-1);
data.y = PM. response;
data.n = ones(size(data.x));
pfhb = PAL_PFHB_fitModel(data,'PF','Gumbel');
plot(priorAlphaRange,PF([PM.threshold(end) 10.^PM.slope(end) PM.guess(end) PM.lapse(end)],priorAlphaRange))
hold on
plot(priorAlphaRange,PF([pfhb.summStats.a.mean 10.^pfhb.summStats.b.mean .5 pfhb.summStats.l.mean],priorAlphaRange))